What, why and how to gate
Even though we generally use sophisticated dimensionality and clustering techniques, you'll still need to gate your data to remove
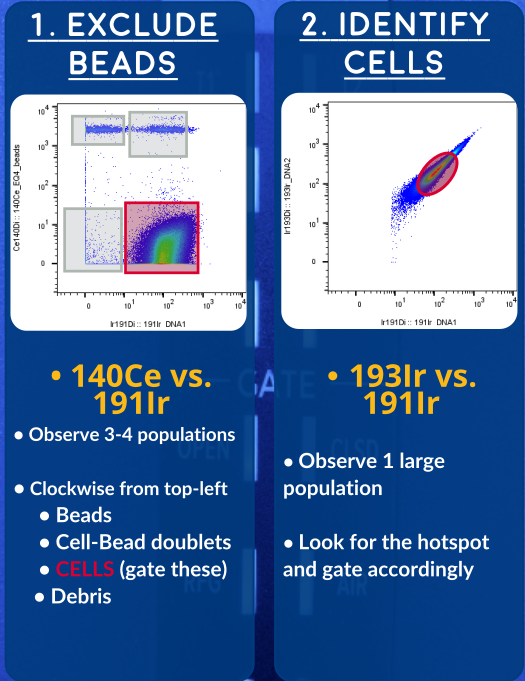
- EQ Beads added to every sample to ensure quality data
- Confounding cell events
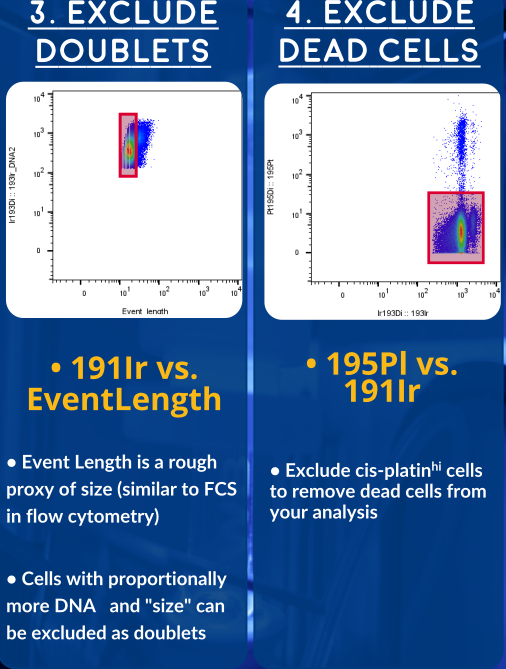
- Dead Cells
- Doublets
- Debris
so that you can provide valid events for downstream algorithms It is also useful to do some manual gating with your first few samples to visualize and understand the data.
Before You Begin
When you receive your mass data files (.fcs) from the Mass Cytometry Service they will have been
- Debarcoded
- If you multiplexed your samples for acquisition we will run the debarcoding algorithm to separate your samples into individual files
- Normalized
- ICP-MS detectors lose sensitivity over the course of a few hours. Data is normalized to account for this using EQ beads added to every sample.
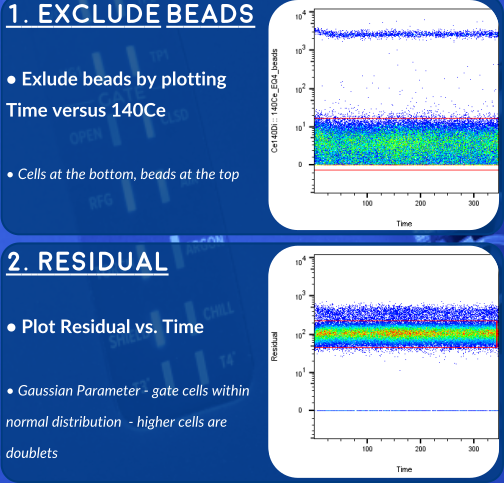
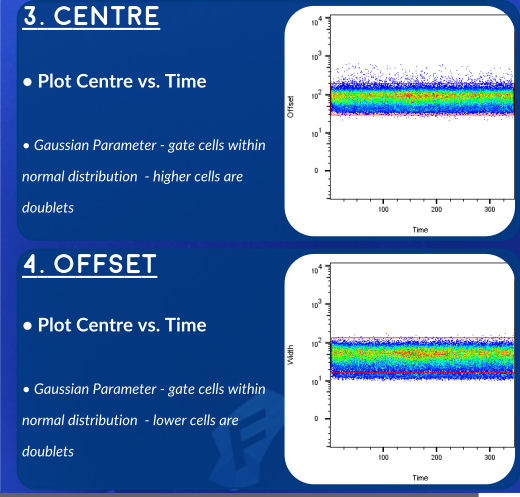
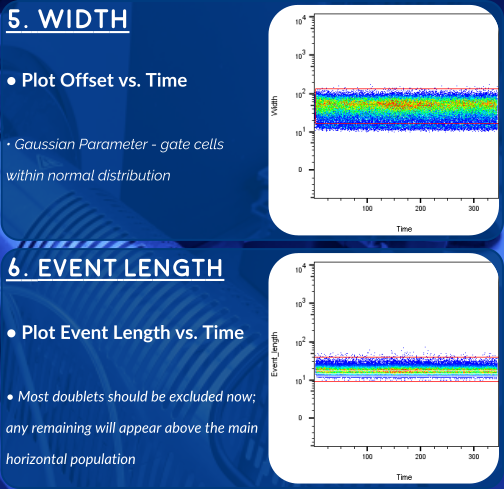
An Alternative Strategy - Gaussian Parameters
The newer generations of CyToF (like the Helios, here at OHRI) allow us to record additional parameters for each event. These gaussian parameters allow us to exclude doublets with even higher stringency. Overleaf you'll find a gating strategy using this newer set of parameters.